Hi there! 👋 I am Jingbo Zhou (周靖博), a Ph.D. student at CAIRI, Westlake University and Zhejiang University, advised by Chair Prof. Stan Z. Li. Before Joining Westlake, I received my B.E. degree of Big Data Science and Technology from Jilin University.
My research interest includes Foundation Model and AI4Science, especially in the Computational Biochemistry. If you are interested, feel free to contact me for future collaborations through Email or WeChat.
🔎 Research Interests
“Man is the measure of all things”
a) Foundation Model
- Graph Foundation Model
- Large Language Models
b) AI for Science
- Computational Biochemistry
- Single Cell Foundation Model
- Artificial Intelligence Virtual Cell
🔥 News
- 2026.01: ✨✨✨ One paper on Graph Transformer & Graph OOD was accepted by WWW 2026.
- 2025.12: 🕶️🕶️🕶️ Invitation to serve as a reviewer for ICML 2026.
- 2025.12: 🕶️🕶️🕶️ Invitation to serve as a reviewer for Transactions on Knowledge Discovery from Data.
- 2025.12: 🕶️🕶️🕶️ Invitation to serve as a reviewer for Pattern Recognition.
- 2025.11: ✨✨✨ Two co-authored paper on denovo peptide sequencing and dna modeling 🔥🔥🔥 oral presentation were accepted by AAAI 2026. Congratulation to @Siyuan!
- 2025.09: 🔥🔥🔥 One co-authored paper on AI for Spectra has been accepted by Nature Methods in principle.
- 2025.09: ✨✨✨ One co-authored paper on single cell pertubation was accepted by NeurIPS 2025
- 2025.09: ⚛️⚛️⚛️ We have open-sourced the hpdex library for high-performance differential gene expression analysis.
- 2025.08: 🕶️🕶️🕶️ Invitation to serve as a reviewer for ICLR 2026 & AAAI 2026.
- 2025.06: 🌴🌴🌴 Have a wonderful time @VALSE 2025 🇨🇳ZHU HAI .
- 2025.04: ✨✨✨ Two co-authored paper on denovo peptide sequencing & single cell pertubation was accepted by IJCAI 2025.
- 2025.04: 🌴🌴🌴 Have a wonderful time @ICLR 2025 🇸🇬Singapore.
- 2025.01: 🕶️🕶️🕶️ Invitation to serve as a reviewer for NeurIPS 2025.
- 2025.01: ✨✨✨ Two co-authored paper on RAG boosting denovo peptide sequencing was accepted by ICLR 2025.
- 2024.12: 🕶️🕶️🕶️ Invitation to serve as a reviewer for ICML 2025.
- 2024.09: ✨✨✨ One co-authored paper on denovo peptide sequencing was accepted by NeurIPS 2024 Workshop AIDrugX (spotlight).
- 2024.10: 🏆🏆🏆 I am awarded NeurIPS Scholar Award.
- 2024.09: 🕶️🕶️🕶️ Invitation to serve as a reviewer for AISTATS 2025.
- 2024.09: ✨✨✨ Two papers on de novo peptide sequencing were accepted by NeurIPS 2024.
- 2024.09: ✨✨✨ One paper on deep GNN was accepted by NeurIPS 2024.
- 2024.08: 🕶️🕶️🕶️ Invitation to serve as a reviewer for ICLR 2025.
- 2024.05: 🕶️🕶️🕶️ Invitation to serve as a reviewer for NeurIPS 2024.
- 2024.06: 🎓🎓🎓 I obtained a Bachelor of Science degree from Jilin University.
- 2023.12: 🏆🏆🏆 I received the Wang Xianghao Scholarship (王湘浩奖学金).
⚛️ Open Source Projects
📃 Publications
† Equal Contribution
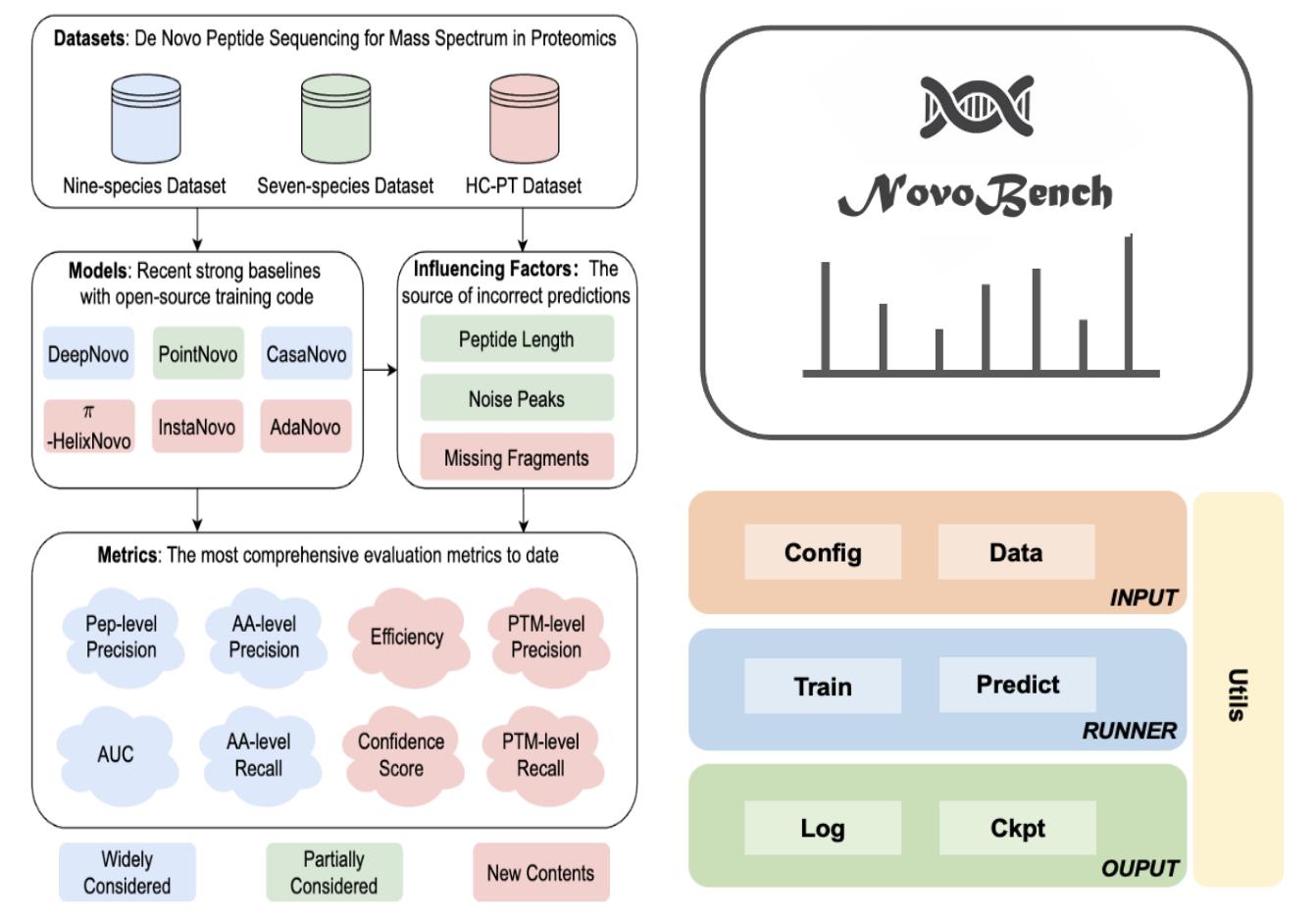
NovoBench: Benchmarking Deep Learning-based De Novo Sequencing Methods in Proteomics
Jingbo Zhou†, Shaorong Chen†, Jun Xia†, Sizhe Liu, Tianze Ling, Wenjie Du, Yue Liu, Jianwei Yin, Stan Z. Li
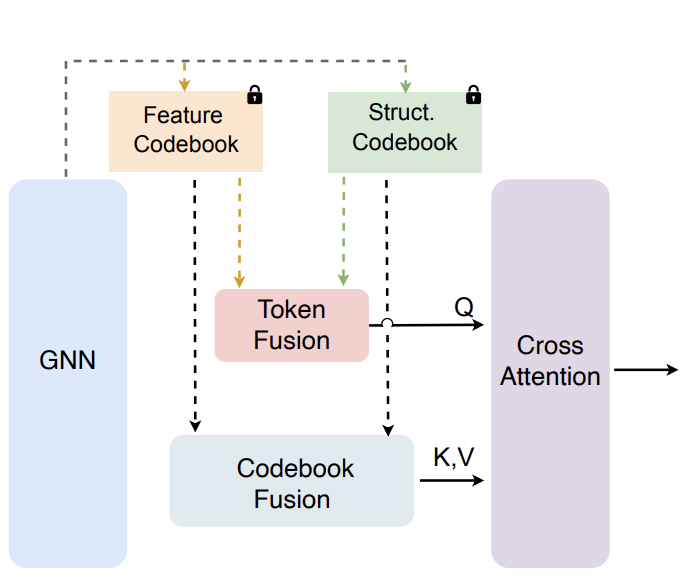
VecFormer: Towards Efficient and Generalizable Graph Transformer with Graph Token Attention
Jingbo Zhou†, Jun Xia†, Chang Yu†, Siyuan Li, Yunfan Liu, Wenjun Wang, Yufei Huang, Changxi Chi, Mutian Hong, Zhuoli Ouyang, Shu Wang, Zhongqi Wang, Xingyu Wu, Stan Z. Li
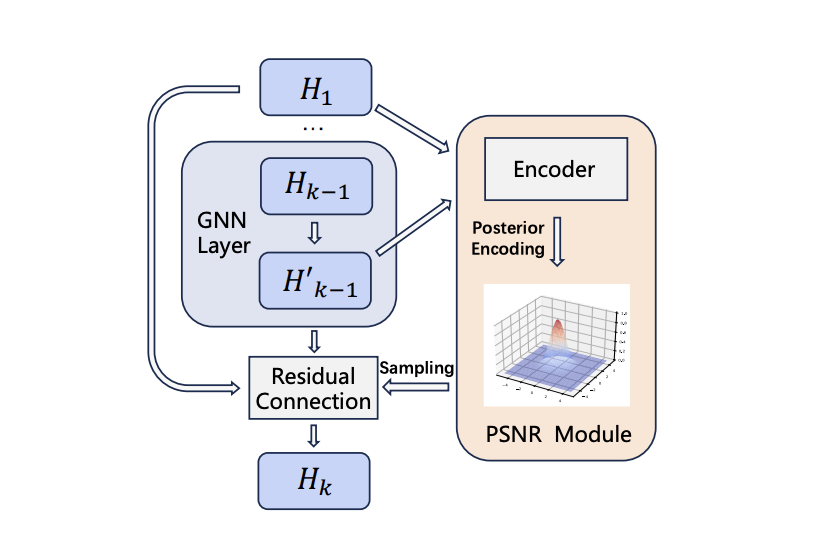
Deep Graph Neural Networks via Posteriori-Sampling-based Node-Adaptative Residual Module
Jingbo Zhou†, Yixuan Du†, Ruqiong Zhang†, Jun Xia, Zhizhi Yu, Zelin Zang, Di Jin, Carl Yang, Rui Zhang, Stan Z. Li
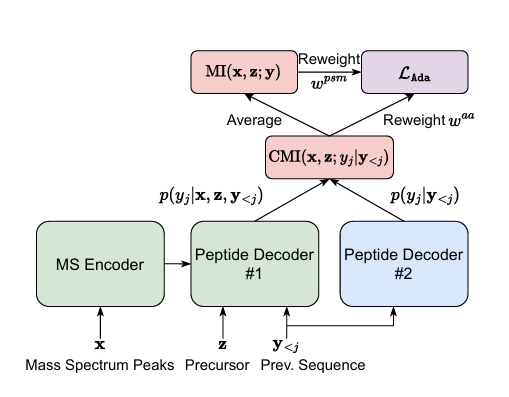
AdaNovo: Adaptive De Novo Peptide Sequencing with Conditional Mutual Information
Jun Xia†, Shaorong Chen†, Jingbo Zhou†, Xiaojun Shan, Wenjie Du, Zhangyang Gao, Cheng Tan, Bozhen Hu, Jiangbin Zheng, Stan Z. Li
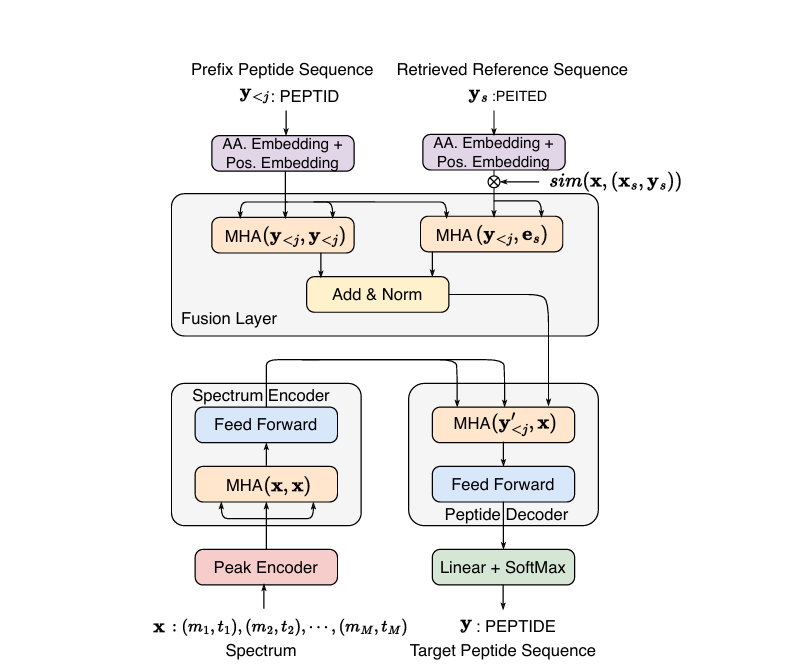
Bridging the Gap between Database Search and De Novo Peptide Sequencing with SearchNovo
Jun Xia†, Sizhe Liu†, Jingbo Zhou†, Shaorong Chen, hongxin xiang, Zicheng Liu, Yue Liu, Stan Z. Li
🔥 NeurIPS 2024 AIDrugX Spotlight
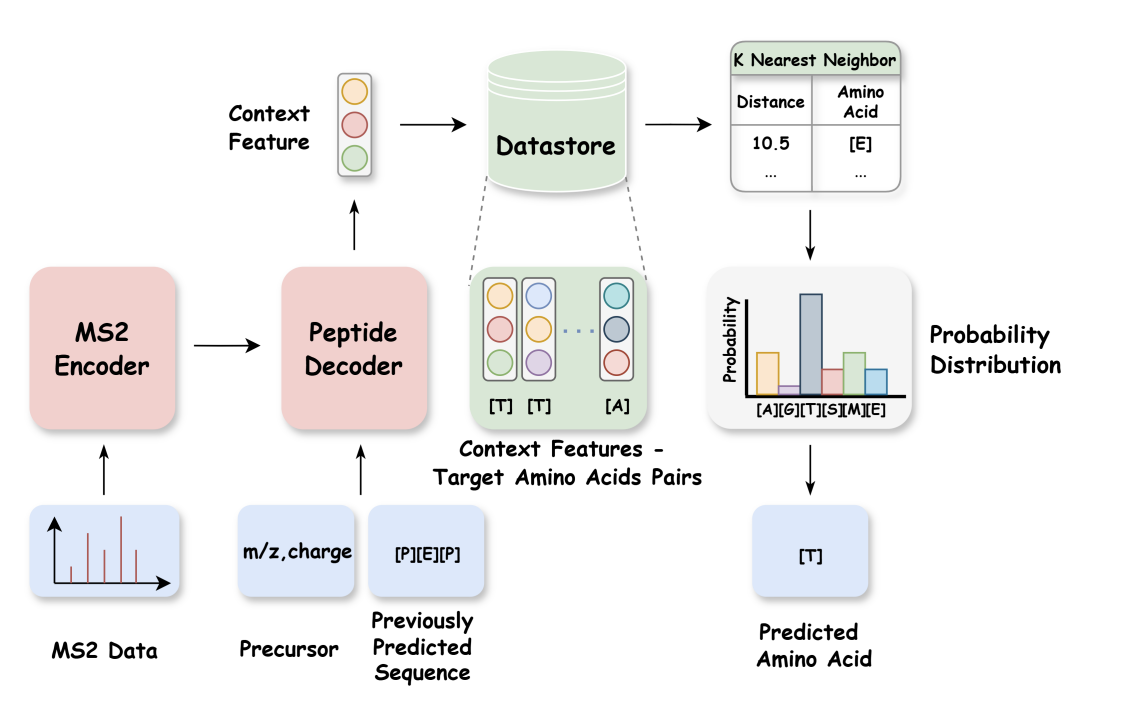
ReNovo: Retrieval-Based De Novo Mass Spectrometry Peptide Sequencing
Shaorong Chen†, Jun Xia†, Jingbo Zhou†, Lecheng Zhang, Zhangyang Gao, Bozhen Hu, Cheng Tan, Wenjie Du, Stan Z. Li
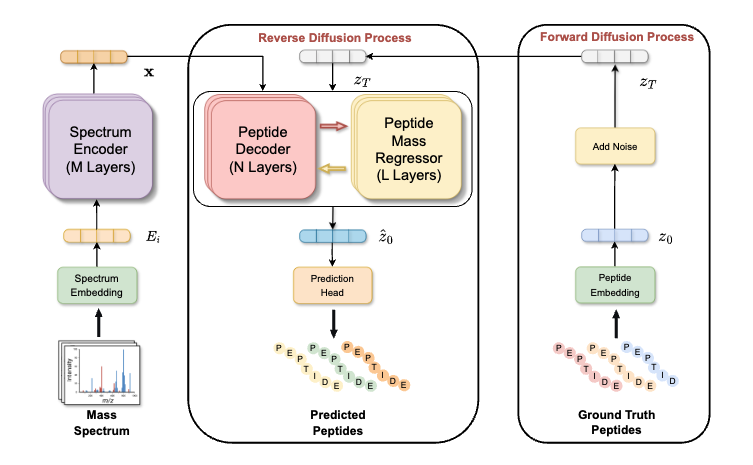
Regressor-guided Diffusion Model for De Novo Peptide Sequencing with Explicit Mass Control
Shaorong Chen, Jingbo Zhou, Jun Xia
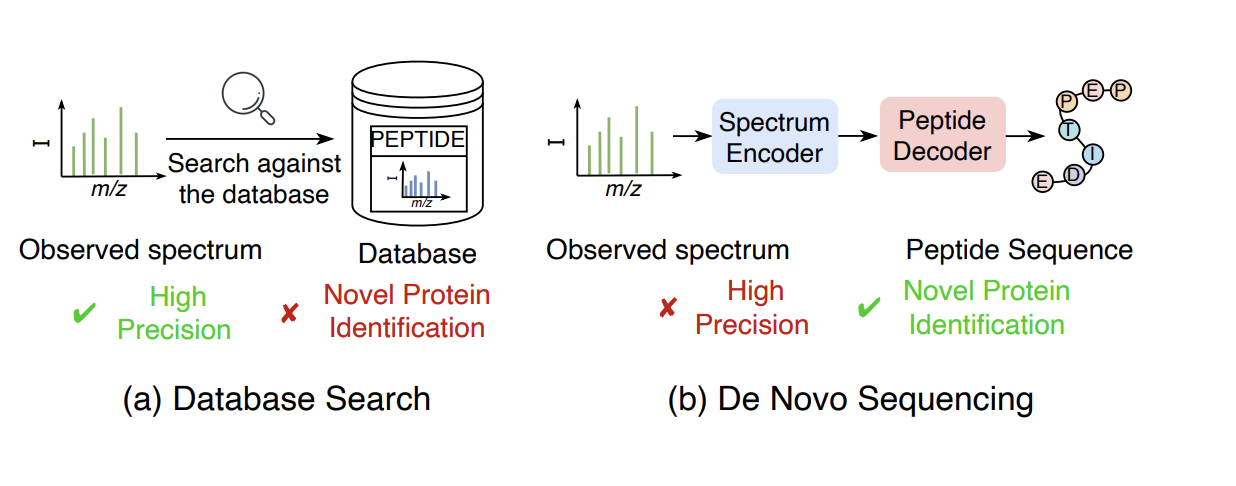
A comprehensive and systematic review for deep learning-based de novo peptide sequencing
Jun Xia†, Jingbo Zhou†, Shaorong Chen†, Tianze Ling†, Stan Z. Li
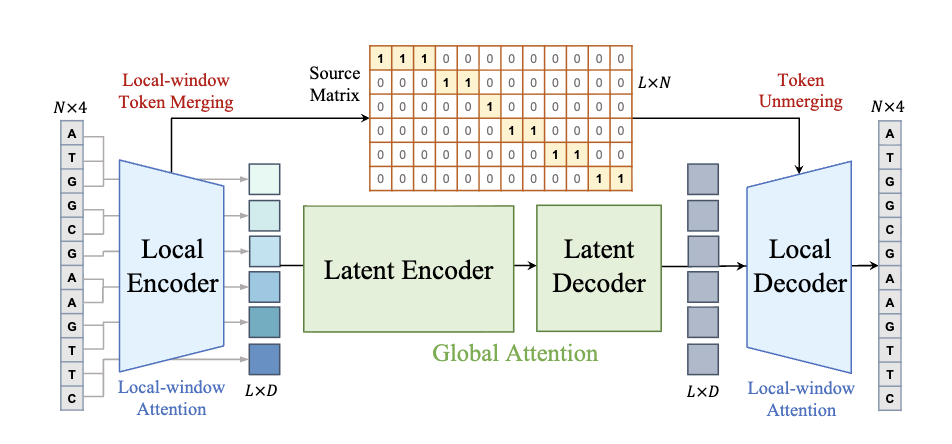
MergeDNA: Context-aware Genome Modeling with Dynamic Tokenization through Token Merging
Siyuan Li, Zicheng Liu, Kai Yu, Chang Yu, Jingbo Zhou, Anna Wang, Qirong Yang, yucheng guo, Xiaoming Zhang, Stan Z. Li
🔥 AAAI 2026 Oral Presentation
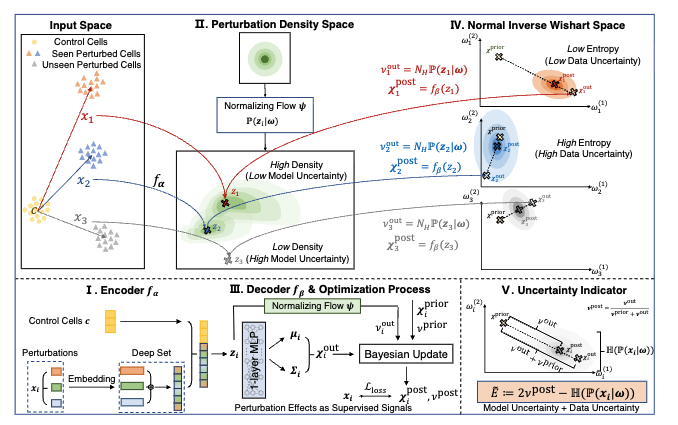
PRESCRIBE: Predicting Single-Cell Responses with Bayesian Estimation
Jiabei Cheng, Changxi Chi, Jingbo Zhou, Hongyi Xin, Jun Xia
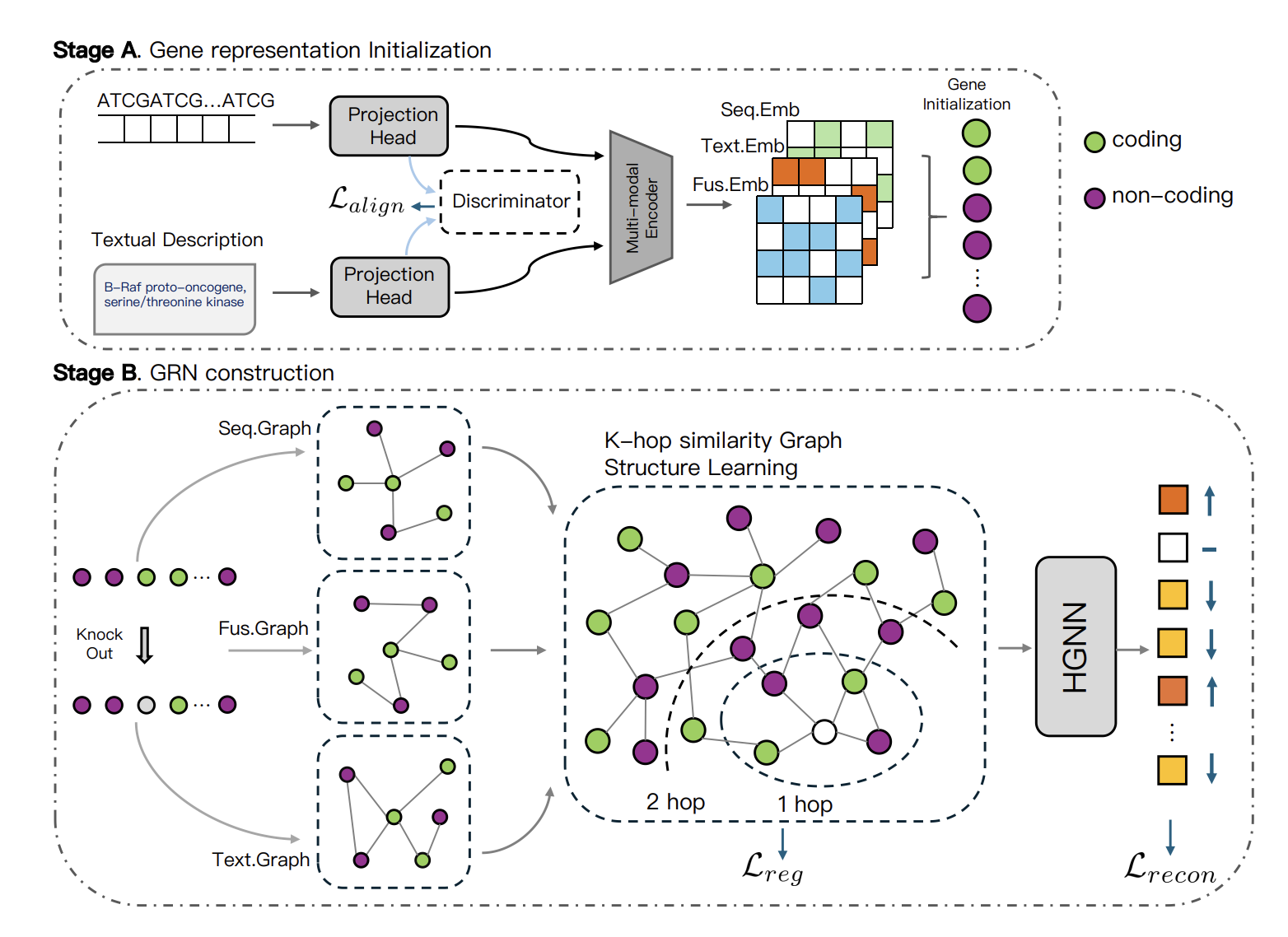
Changxi Chi, Xia Jun, Jingbo Zhou, Jiabei Cheng, Chang Yu, Stan Z. Li
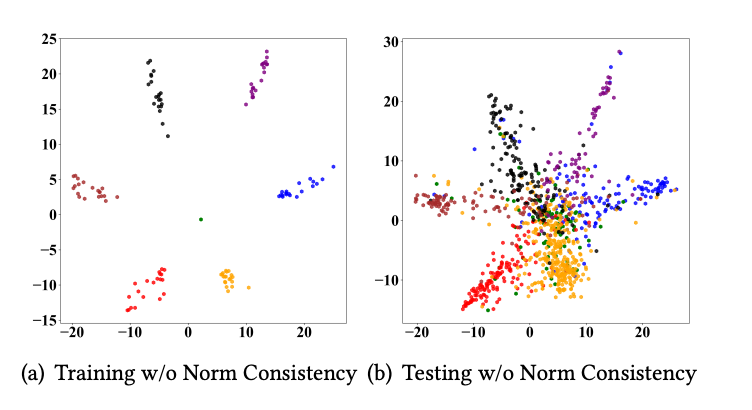
Shenzhi Yang, Jun Xia, Jingbo Zhou, Xingkai Yao, Xiaofang Zhang
📖 Education
Ph.D., Engineering College, Westlake University & Zhejiang University, Hangzhou


Undergraduate, Software College, Jilin University, Changchun

💬 Service
Conference Reviewer
- Reviewer for NeurIPS’2024, 2025
- Reviewer for ICML’2025, 2026
- Reviewer for ICLR’2025, 2026
- Reviewer for AISTATS’2025, 2026
- Reviewer for AAAI’2025
Journal Reviewer
- Reviewer for Pattern Recognition
- Reviewer for TKDD
- Reviewer for TCBB

